Covid 19 - Mapping Cases - Santiago
After the arrival of Covid 19 to Chile on the 3rd March 2020, the virus has spread throughout the country. Nonetheless the capital - Santiago and the Metropolitan Region in which it resides, have been by far the hardest hit location with 254,354 registered cases in this Region as of the 29th July 2020. This publication focuses on mapping the data for confirmed cases in Santiago and the Metropolitan Region.
2) Packages
The following packages are used in this publication.
library(chilemapas)
library(ggplot2)
library(stringr)
library(tidyverse)
library(ggspatial)3) Creating a Base Map
The package chilemapas is used to create a base map for the Metropolitan Region, in which Santiago resides. Additionally, accents are added to the names of each of the comunas, so that they can be combined with other data bases that use accents in their spelling of the comunas.
comunas <- chilemapas::mapa_comunas
codigos <- chilemapas::codigos_territoriales
comunas_info <- left_join(codigos, comunas)
comunas_info_RM <- comunas_info %>% filter(nombre_region == 'Metropolitana de Santiago')
comunas_info_RM[4, 2] = "Conchalí"
comunas_info_RM[6, 2] = "Estación Central"
comunas_info_RM[19, 2] = "Maipú"
comunas_info_RM[20, 2] = "Ñuñoa"
comunas_info_RM[22, 2] = "Peñalolén"
comunas_info_RM[29, 2] = "San Joaquín"
comunas_info_RM[31, 2] = "San Ramón"
ggplot() + geom_sf(data = comunas_info_RM$geometry) + ggtitle("Base Map of the Metropolitan Region") + ylab("Latitude") + xlab("Longitude")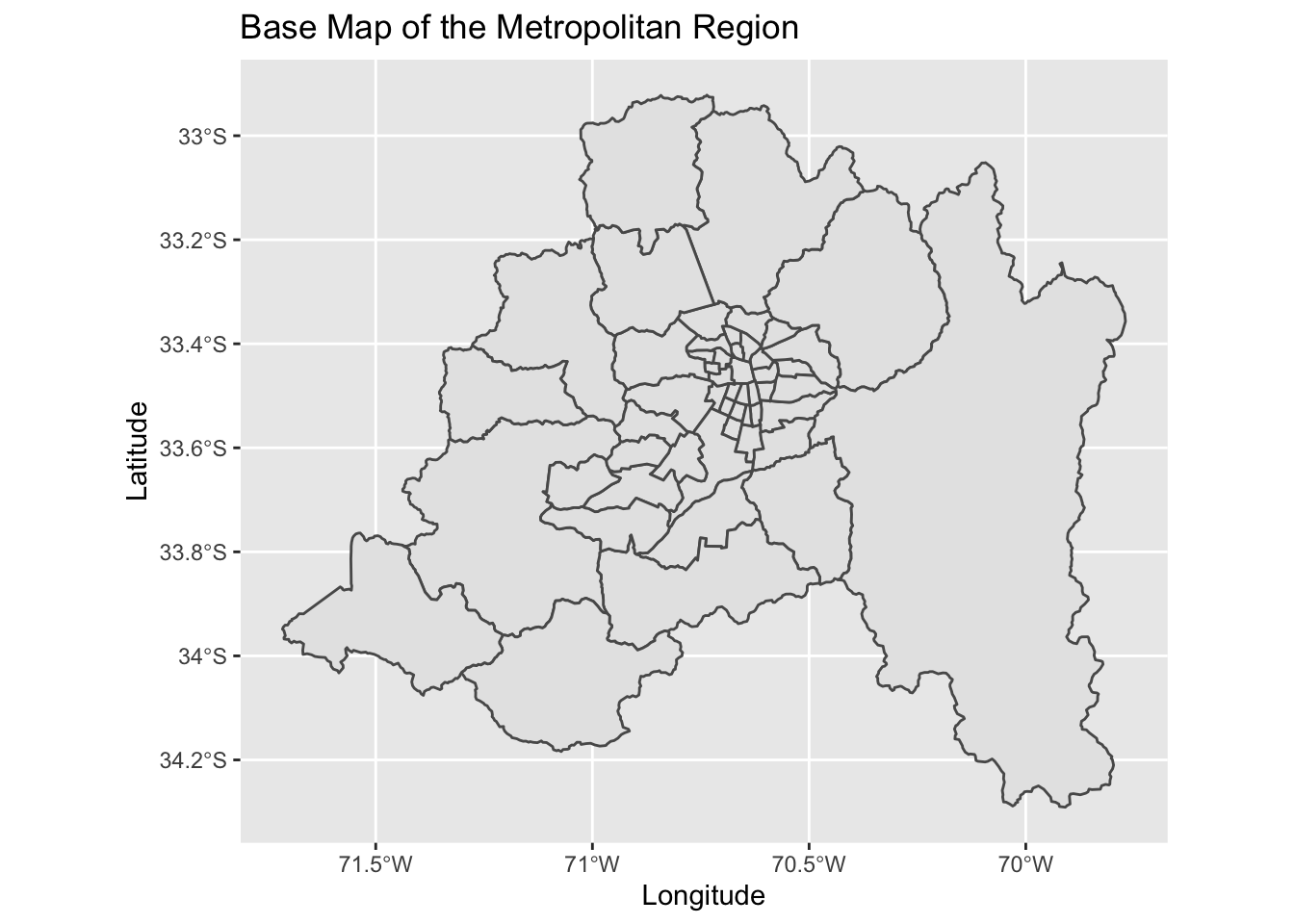
4) Data for Known Cases
The data for accumulated cases can be downloaded from the github of the Ministry of Health. With the data already downloaded it is loaded using the following syntax.
confirmedcases <- read_csv("~/Documents/Machine Learning/15. Hugo/academic-kickstart-master/content/en/post/MappingCovid/Covid-19_T.csv")5) Feature Engineering
The objective of this section is to prepare the data so that it can be combined with the base map data in comunas_info_RM.
Firstly, the data is filtered so as to only include the comunas in the Metropolitan Region. Then the names in row two are made the variable names. Then rows one to four are removed.
The variables are converted to numerical variables.
RM %>% mutate_all(type.convert) %>%
mutate_if(is.character, as.numeric) %>%
mutate_if(is.factor, as.Date)## # A tibble: 37 x 54
## Comuna Alhue Buin `Calera de Tang… Cerrillos `Cerro Navia` Colina
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020-03-30 0 7 6 0 19 32
## 2 2020-04-01 0 8 7 4 21 39
## 3 2020-04-03 0 14 7 4 26 47
## 4 2020-04-06 0 20 7 5 31 50
## 5 2020-04-08 0 20 8 11 36 62
## 6 2020-04-10 0 29 8 21 42 66
## 7 2020-04-13 0 36 10 29 57 74
## 8 2020-04-15 0 40 10 39 65 77
## 9 2020-04-17 0 43 11 48 79 81
## 10 2020-04-20 0 47 14 53 99 90
## # … with 27 more rows, and 47 more variables: Conchali <dbl>, Curacavi <dbl>,
## # `El Bosque` <dbl>, `El Monte` <dbl>, `Estacion Central` <dbl>,
## # Huechuraba <dbl>, Independencia <dbl>, `Isla de Maipo` <dbl>, `La
## # Cisterna` <dbl>, `La Florida` <dbl>, `La Granja` <dbl>, `La Pintana` <dbl>,
## # `La Reina` <dbl>, Lampa <dbl>, `Las Condes` <dbl>, `Lo Barnechea` <dbl>,
## # `Lo Espejo` <dbl>, `Lo Prado` <dbl>, Macul <dbl>, Maipu <dbl>, `Maria
## # Pinto` <dbl>, Melipilla <dbl>, Nunoa <dbl>, `Padre Hurtado` <dbl>,
## # Paine <dbl>, `Pedro Aguirre Cerda` <dbl>, Penaflor <dbl>, Penalolen <dbl>,
## # Pirque <dbl>, Providencia <dbl>, Pudahuel <dbl>, `Puente Alto` <dbl>,
## # Quilicura <dbl>, `Quinta Normal` <dbl>, Recoleta <dbl>, Renca <dbl>, `San
## # Bernardo` <dbl>, `San Joaquin` <dbl>, `San Jose de Maipo` <dbl>, `San
## # Miguel` <dbl>, `San Pedro` <dbl>, `San Ramon` <dbl>, Santiago <dbl>,
## # Talagante <dbl>, Tiltil <dbl>, Vitacura <dbl>, `Desconocido
## # Metropolitana` <dbl>The data is filtered to only include two dates for each month, one on the 1st and one on the 15th. The data base is also converted from a wide format to a long format. Then the data base is divided by the date so as to have individual variables for each of the required dates. This process leaves the data in a matrix with a transformation to a data frame format being required. Additionally, here the excess date variables are deleted leaving only a variable for comuna and variables for the different dates. Then the variables are renamed and those which are numerical are converted to factors. Finally, row 9, which refers to cases with an unknown comuna, is removed.
RM_fechas <- RM %>% filter(Comuna %in% c('2020-03-30', '2020-04-15', '2020-05-01', '2020-05-15', '2020-06-01', '2020-06-15', '2020-07-01',
'2020-07-13', '2020-07-24'))
RM_long <- RM_fechas %>%
gather(key = "fecha",
value = "Casos_Confirmados",
c(-Comuna))
RM_long %>% arrange(Comuna)## # A tibble: 477 x 3
## Comuna fecha Casos_Confirmados
## <chr> <chr> <chr>
## 1 2020-03-30 Alhue 0.0
## 2 2020-03-30 Buin 7.0
## 3 2020-03-30 Calera de Tango 6.0
## 4 2020-03-30 Cerrillos 0.0
## 5 2020-03-30 Cerro Navia 19.0
## 6 2020-03-30 Colina 32.0
## 7 2020-03-30 Conchali 14.0
## 8 2020-03-30 Curacavi 5.0
## 9 2020-03-30 El Bosque 18.0
## 10 2020-03-30 El Monte 0.0
## # … with 467 more rowsRM_long <- split(RM_long, RM_long$fecha)
class(RM_long)## [1] "list"RM_long <- data.frame(matrix(unlist(RM_long), nrow=length(RM_long), byrow=T))
RM_long <- RM_long[,18:27]
colnames(RM_long)[1:10] <- c("Comuna", '2020-03-30', '2020-04-15', '2020-05-01', '2020-05-15', '2020-06-01', '2020-06-15', '2020-07-01', '2020-07-13', '2020-07-24')
RM_long <- cbind(RM_long[,1], RM_long[,-1] %>% mutate_all(type.convert) %>%
mutate_if(is.factor, as.numeric))
colnames(RM_long)[1] <- "Comuna"
RM_long <- RM_long[-9,]Now the data for cases are combined with the base map data.
casos_en_comunas <- cbind(comunas_info_RM %>% arrange(nombre_comuna), RM_long)6) Maps
Below there are varios maps showing how Covid 19 has spread in the Metropolitan Region. The data is for accumulated cases. Thank you for reading this publication and hopefully the maps are of interest and useful.
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-03-30`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 30 Marzo 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")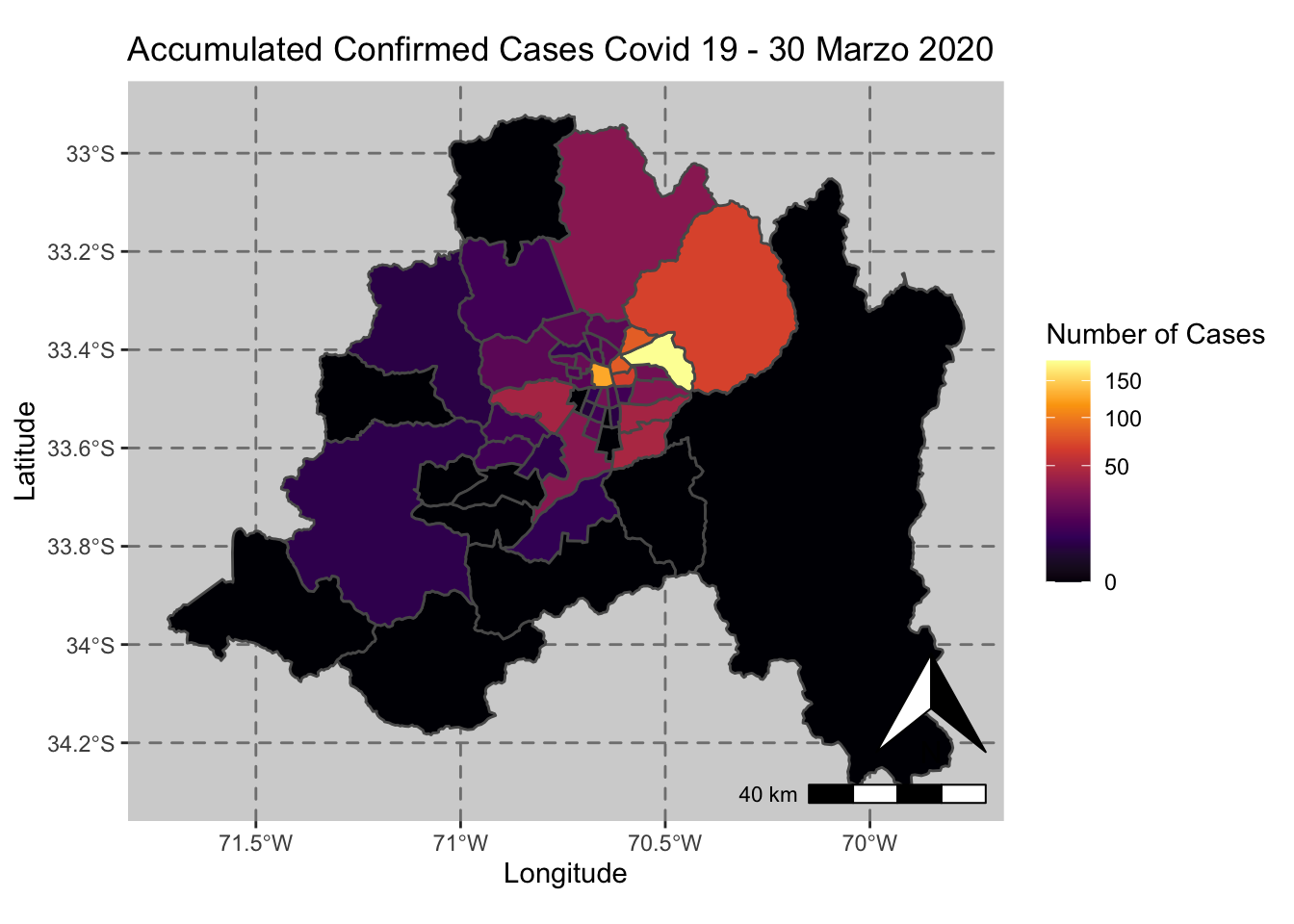
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-04-15`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 15 Abril 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")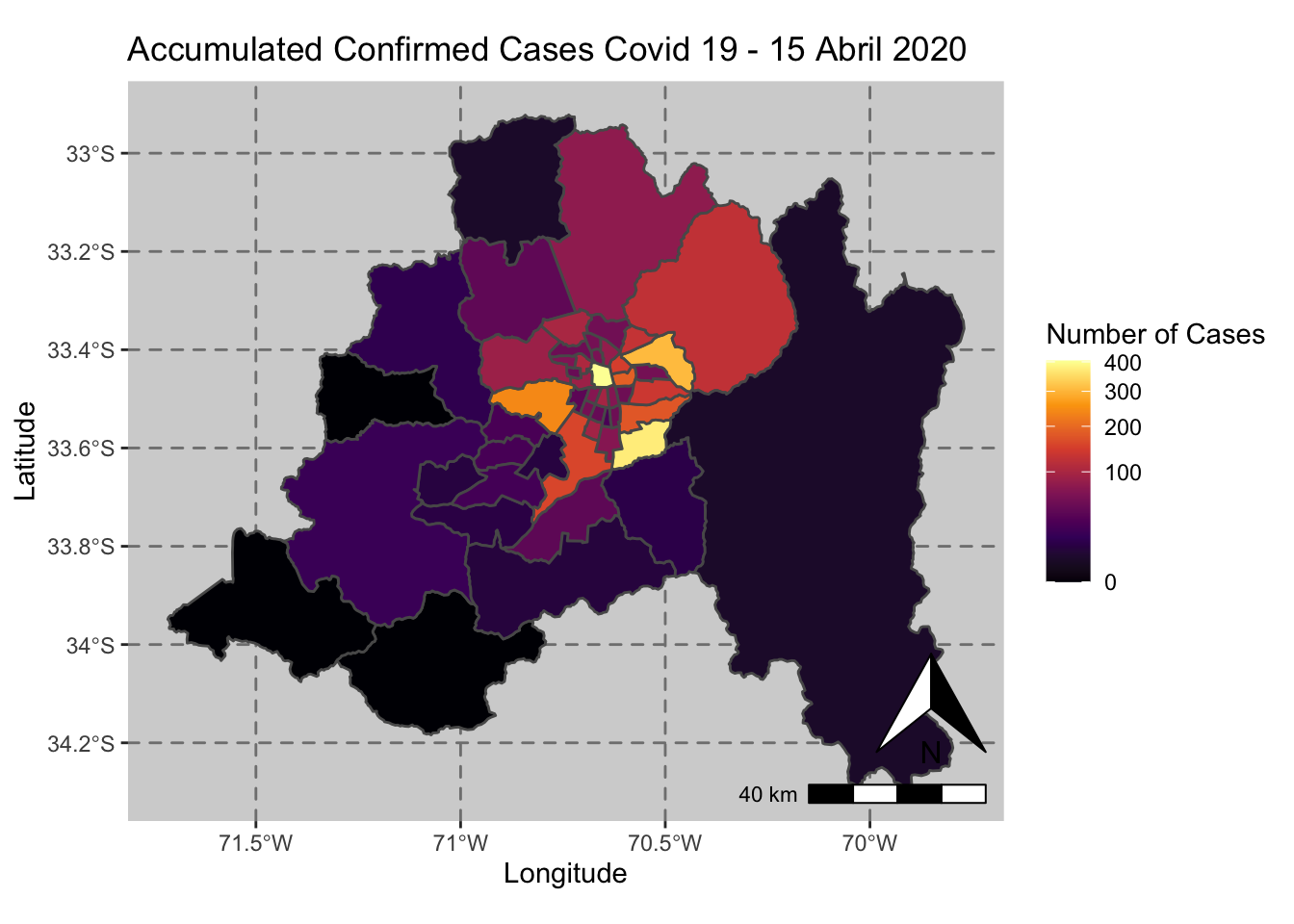
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-05-01`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 01 Mayo 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")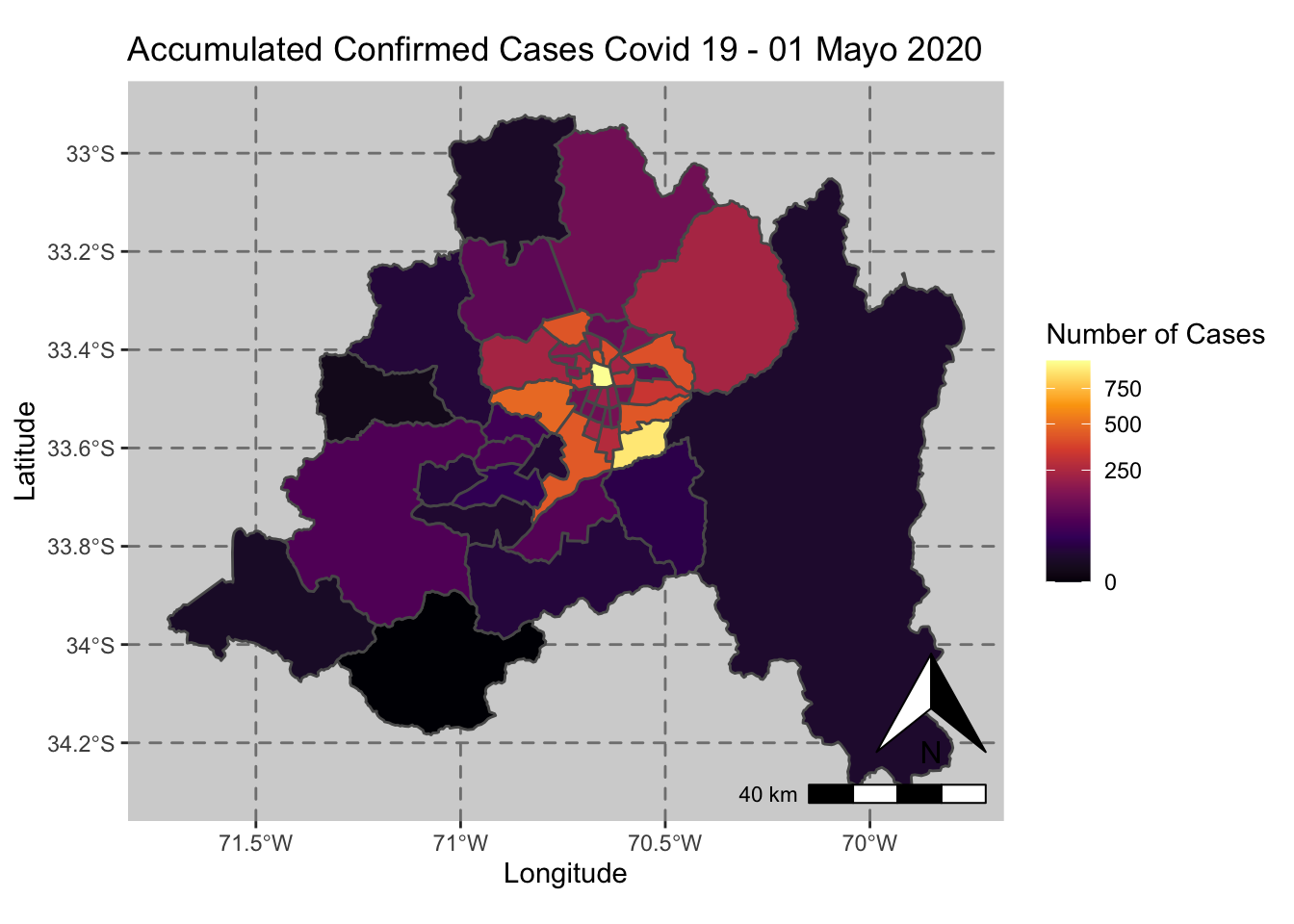
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-05-15`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 15 Mayo 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")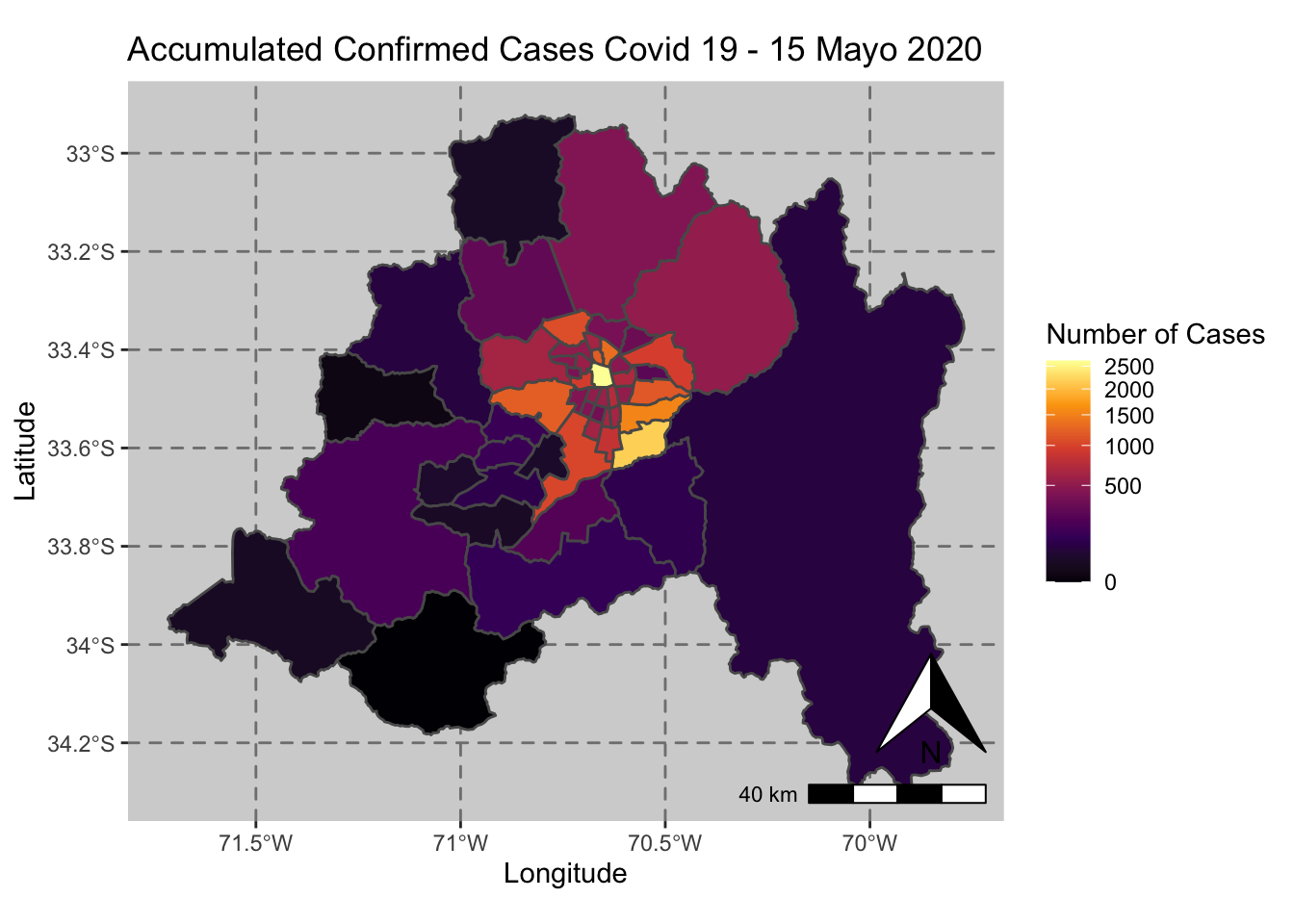
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-06-01`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 01 Junio 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")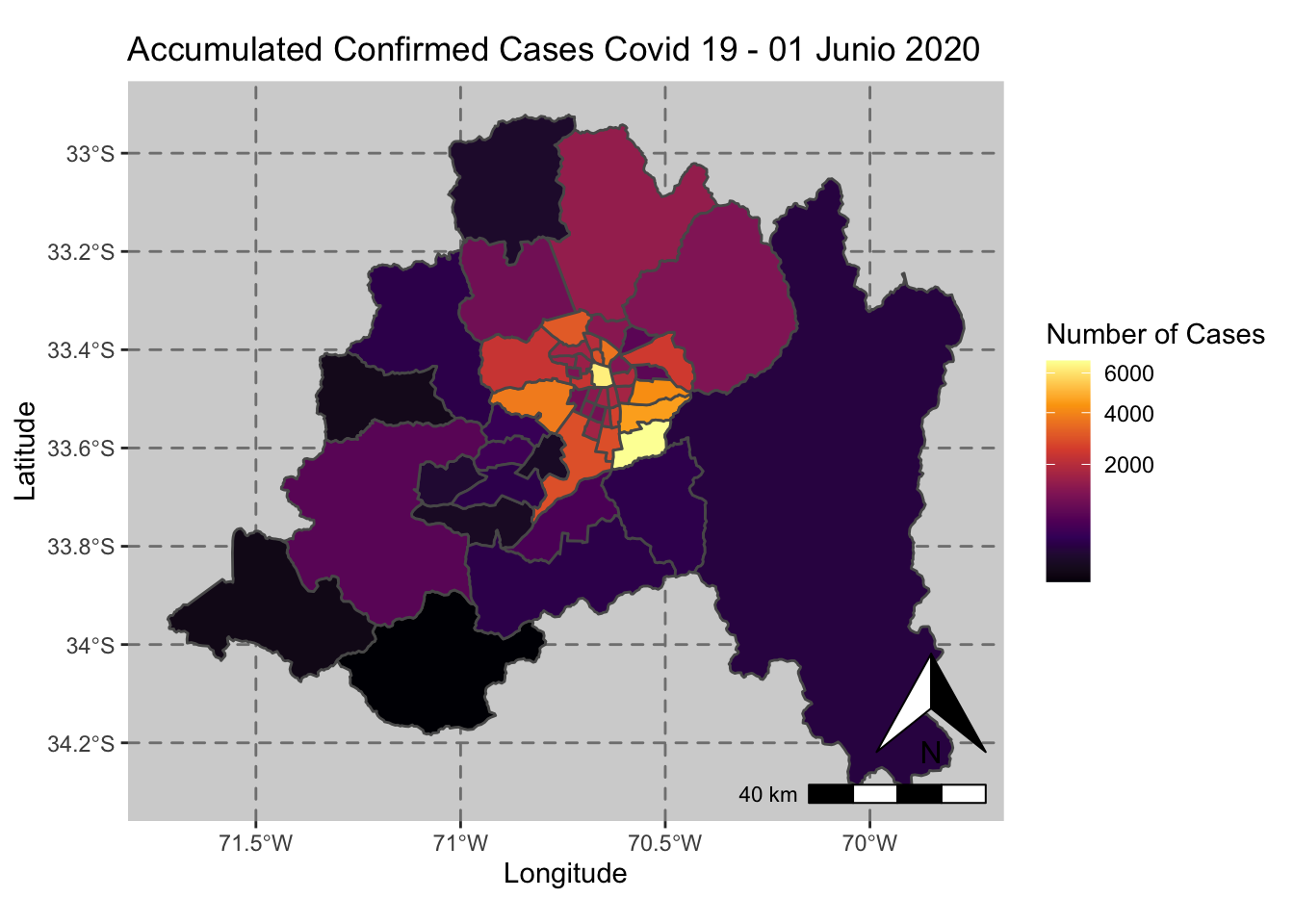
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-06-15`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 15 Junio 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")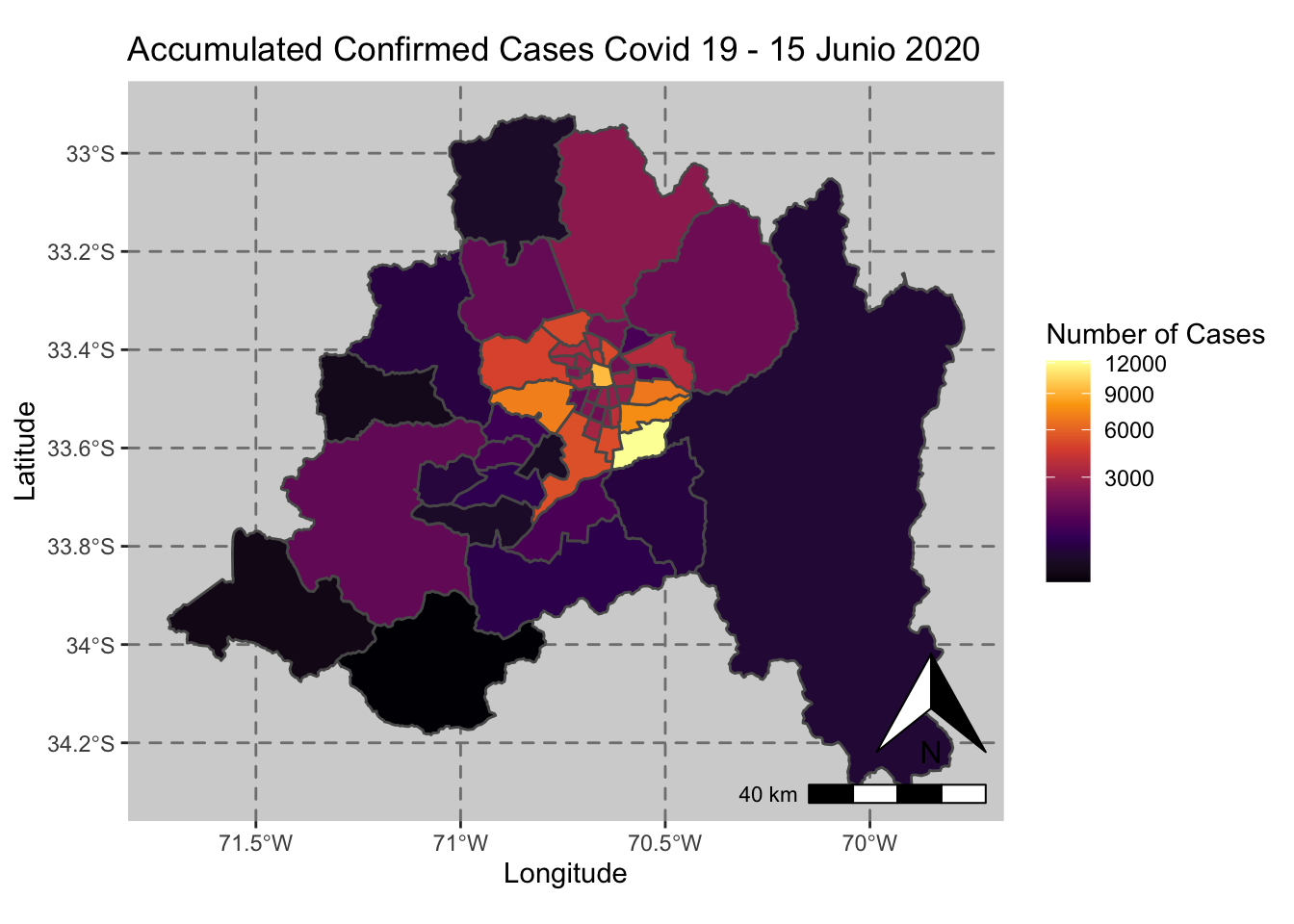
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-07-01`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 01 Julio 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")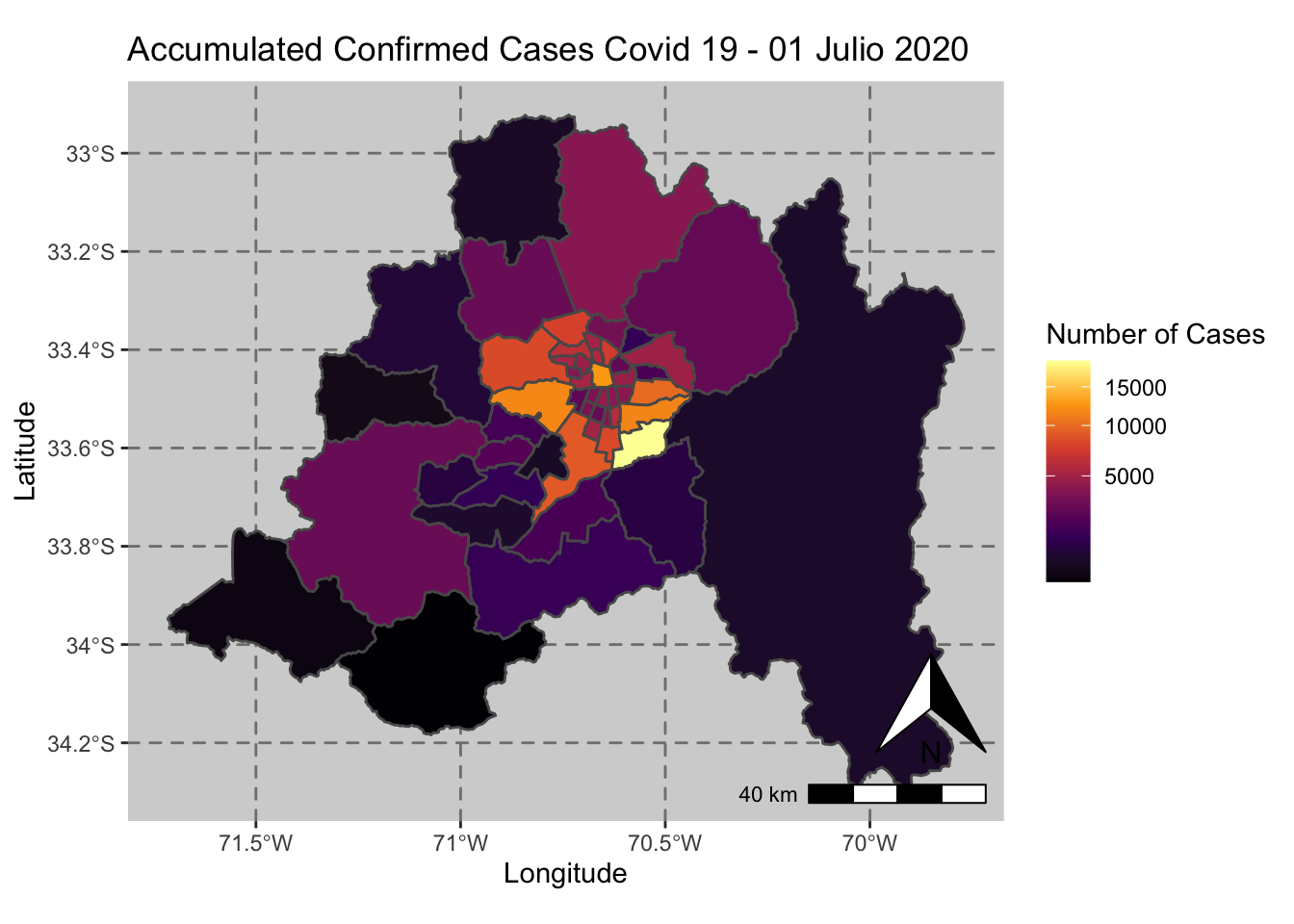
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-07-13`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 13 Julio 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")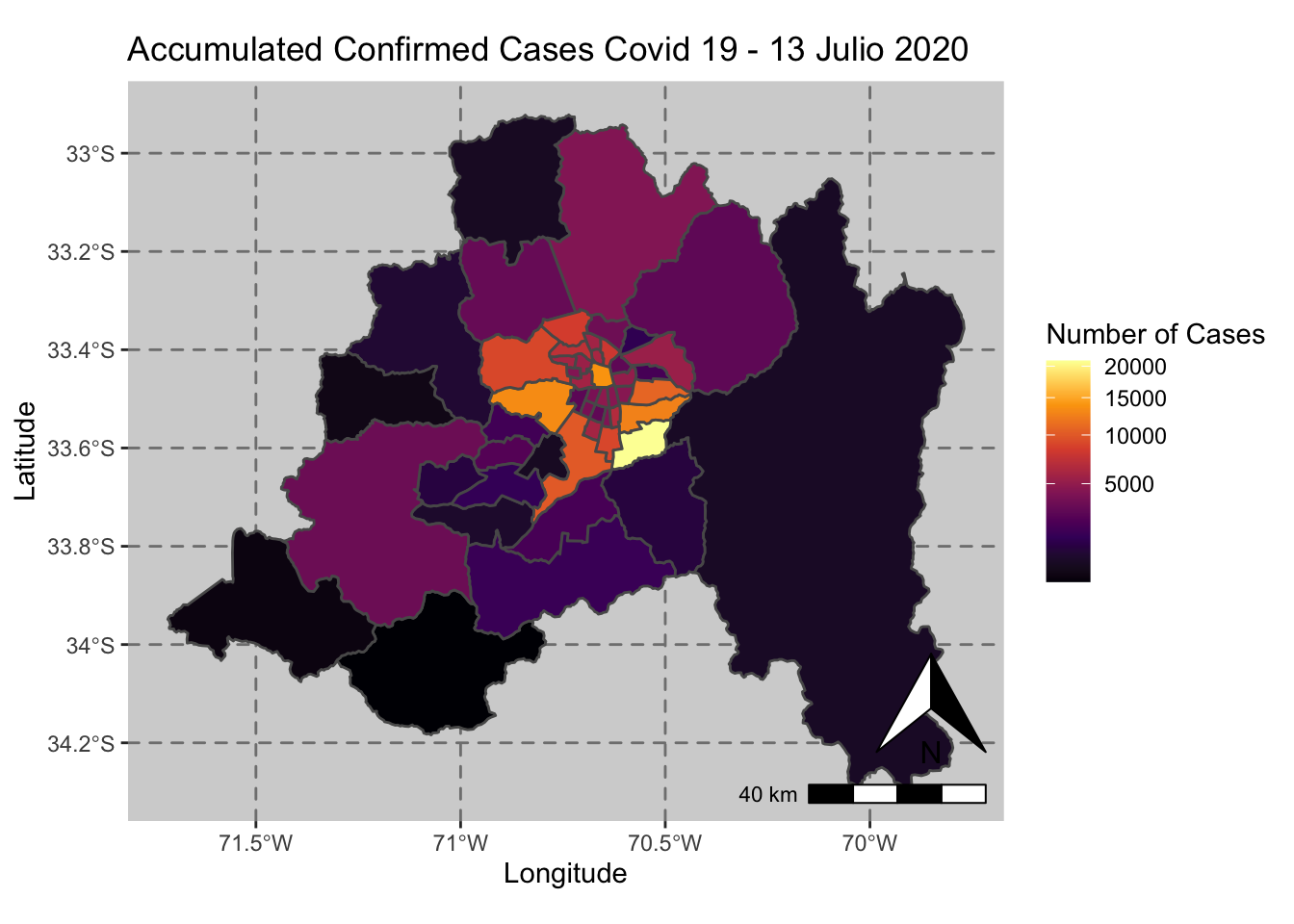
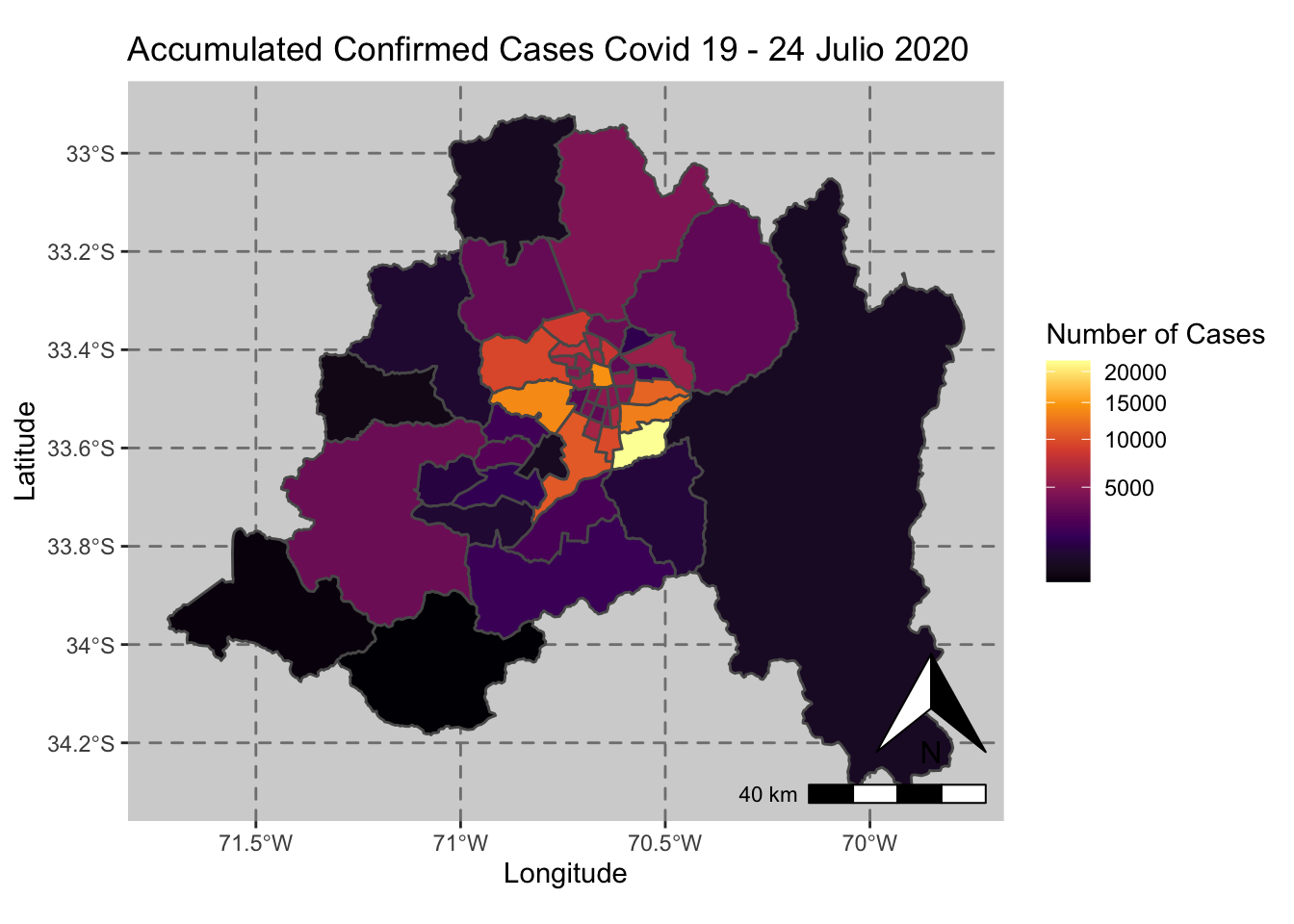
ggplot() + geom_sf(data = casos_en_comunas$geometry, aes(fill = casos_en_comunas$`2020-07-24`)) +
scale_fill_viridis_c(option = "inferno",trans = 'sqrt') +
annotation_north_arrow(aes(which_north = "true", location = "br"), pad_y = unit(0.8, "cm")) +
annotation_scale(aes(location = "br", style = "bar")) +
theme(panel.grid.major = element_line(color = gray(0.5), linetype = "dashed")) +
theme (panel.background = element_rect(fill = "light grey")) +
ggtitle("Accumulated Confirmed Cases Covid 19 - 24 Julio 2020 ") + xlab("Longitude") + ylab("Latitude") +
labs(fill = "Number of Cases")